Two graphical objects are obtained, one corresponding to row labeling and the second to the table of
numbers of patients at risk. If title = TRUE, a third object corresponding to the table title is
also obtained.
Arguments
- data
(
data.frame)
survival data as pre-processed byh_data_plot.- annot_tbl
(
data.frame)
annotation as prepared bysurvival::summary.survfit()which includes the number of patients at risk at given time points.- xlim
(
numeric)
the maximum value on the x-axis (used to ensure the at risk table aligns with the KM graph).- title
(
flag)
whether the "Patients at Risk" title should be added above theannot_at_risktable. Has no effect ifannot_at_riskisFALSE. Defaults toTRUE.
Value
A named list of two gTree objects if title = FALSE: at_risk and label, or three
gTree objects if title = TRUE: at_risk, label, and title.
Examples
# \donttest{
library(dplyr)
library(survival)
library(grid)
fit_km <- tern_ex_adtte %>%
filter(PARAMCD == "OS") %>%
survfit(form = Surv(AVAL, 1 - CNSR) ~ ARMCD, data = .)
data_plot <- h_data_plot(fit_km = fit_km)
xticks <- h_xticks(data = data_plot)
gg <- h_ggkm(
data = data_plot,
censor_show = TRUE,
xticks = xticks, xlab = "Days", ylab = "Survival Probability",
title = "tt", footnotes = "ff", yval = "Survival"
)
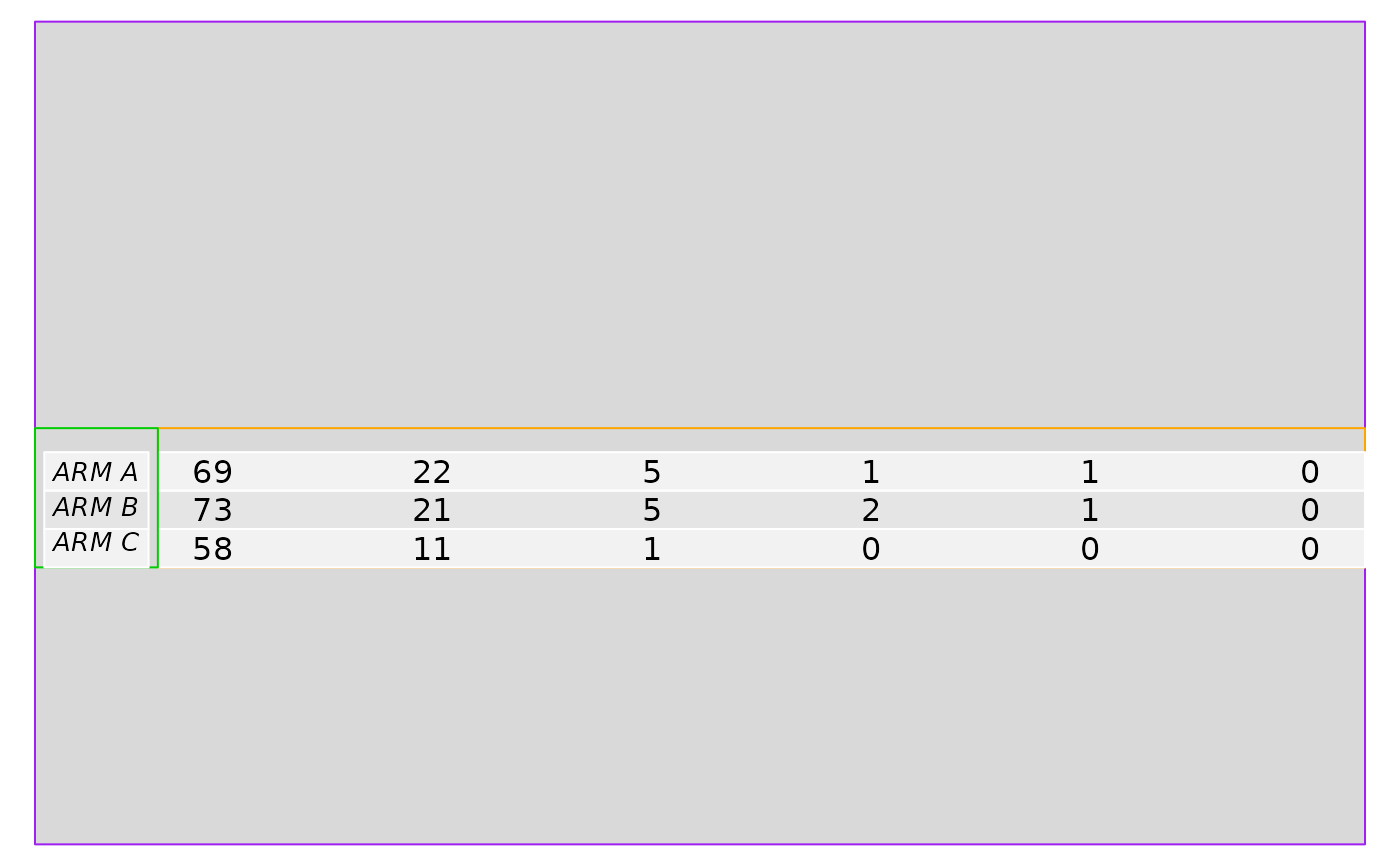
# The annotation table reports the patient at risk for a given strata and
# time (`xticks`).
annot_tbl <- summary(fit_km, time = xticks)
if (is.null(fit_km$strata)) {
annot_tbl <- with(annot_tbl, data.frame(n.risk = n.risk, time = time, strata = "All"))
} else {
strata_lst <- strsplit(sub("=", "equals", levels(annot_tbl$strata)), "equals")
levels(annot_tbl$strata) <- matrix(unlist(strata_lst), ncol = 2, byrow = TRUE)[, 2]
annot_tbl <- data.frame(
n.risk = annot_tbl$n.risk,
time = annot_tbl$time,
strata = annot_tbl$strata
)
}
# The annotation table is transformed into a grob.
tbl <- h_grob_tbl_at_risk(data = data_plot, annot_tbl = annot_tbl, xlim = max(xticks))
# For the representation, the layout is estimated for which the decomposition
# of the graphic element is necessary.
g_el <- h_decompose_gg(gg)
lyt <- h_km_layout(data = data_plot, g_el = g_el, title = "t", footnotes = "f")
grid::grid.newpage()
pushViewport(viewport(layout = lyt, height = .95, width = .95))
grid.rect(gp = grid::gpar(lty = 1, col = "purple", fill = "gray85", lwd = 1))
pushViewport(viewport(layout.pos.row = 3:4, layout.pos.col = 2))
grid.rect(gp = grid::gpar(lty = 1, col = "orange", fill = "gray85", lwd = 1))
grid::grid.draw(tbl$at_risk)
popViewport()
pushViewport(viewport(layout.pos.row = 3:4, layout.pos.col = 1))
grid.rect(gp = grid::gpar(lty = 1, col = "green3", fill = "gray85", lwd = 1))
grid::grid.draw(tbl$label)
 # }
# }