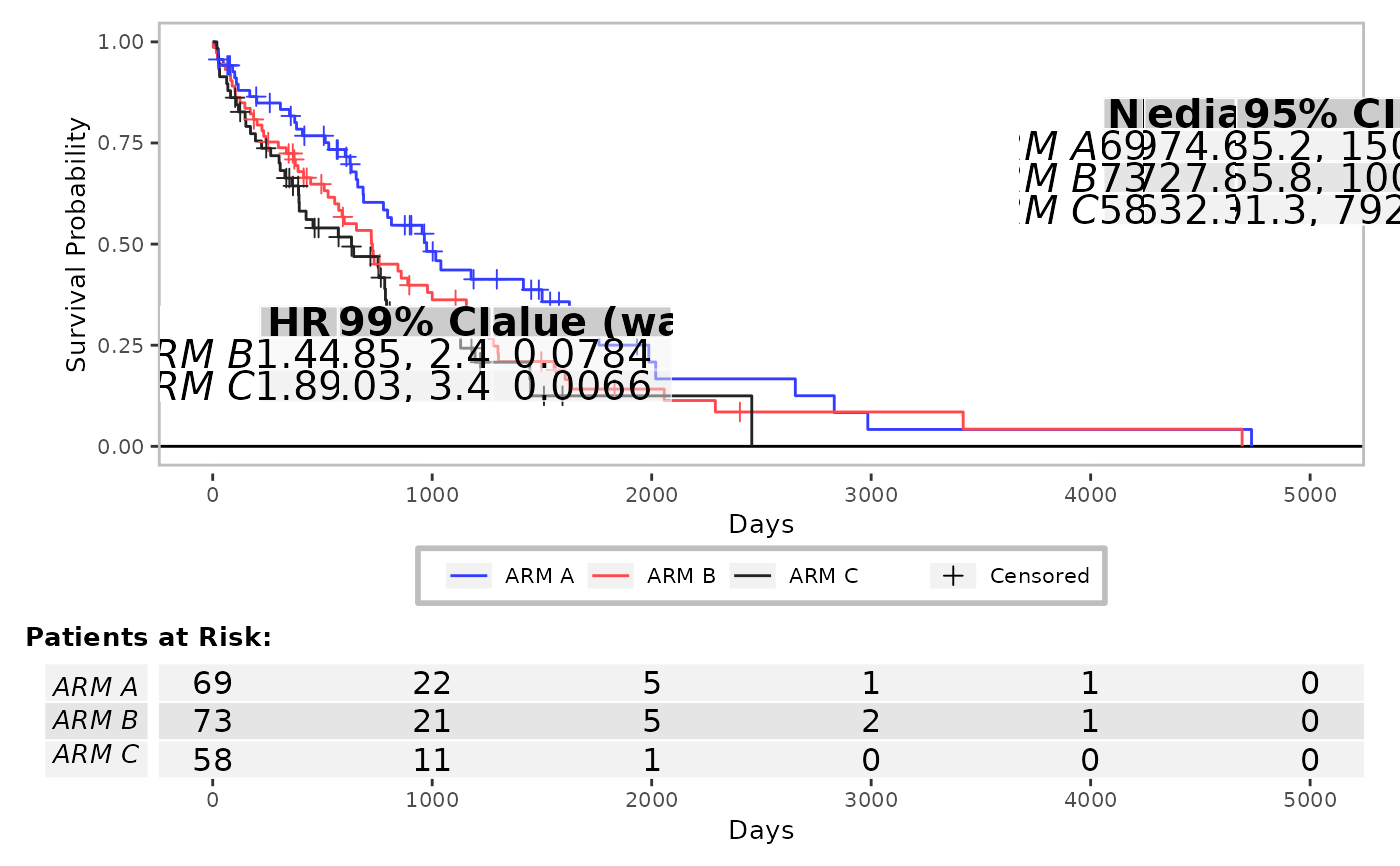
From a survival model, a graphic is rendered along with tabulated annotation including the number of patient at risk at given time and the median survival per group.
Usage
g_km(
df,
variables,
control_surv = control_surv_timepoint(),
col = NULL,
lty = NULL,
lwd = 0.5,
censor_show = TRUE,
pch = 3,
size = 2,
max_time = NULL,
xticks = NULL,
xlab = "Days",
yval = c("Survival", "Failure"),
ylab = paste(yval, "Probability"),
ylim = NULL,
title = NULL,
footnotes = NULL,
draw = TRUE,
newpage = TRUE,
gp = NULL,
vp = NULL,
name = NULL,
font_size = 12,
ci_ribbon = FALSE,
ggtheme = nestcolor::theme_nest(),
annot_at_risk = TRUE,
annot_at_risk_title = TRUE,
annot_surv_med = TRUE,
annot_coxph = FALSE,
annot_stats = NULL,
annot_stats_vlines = FALSE,
control_coxph_pw = control_coxph(),
position_coxph = c(-0.03, -0.02),
position_surv_med = c(0.95, 0.9),
width_annots = list(surv_med = grid::unit(0.3, "npc"), coxph = grid::unit(0.4, "npc"))
)Arguments
- df
(
data.frame)
data set containing all analysis variables.- variables
-
(named
list)
variable names. Details are:tte(numeric)
variable indicating time-to-event duration values.is_event(logical)
event variable.TRUEif event,FALSEif time to event is censored.arm(factor)
the treatment group variable.strat(characterorNULL)
variable names indicating stratification factors.
- control_surv
-
(
list)
parameters for comparison details, specified by using the helper functioncontrol_surv_timepoint(). Some possible parameter options are:conf_level(proportion)
confidence level of the interval for survival rate.conf_type(string)"plain"(default),"log","log-log"for confidence interval type, see more insurvival::survfit(). Note that the option "none" is no longer supported.
- col
(
character)
lines colors. Length of a vector should be equal to number of strata fromsurvival::survfit().- lty
(
numeric)
line type. Length of a vector should be equal to number of strata fromsurvival::survfit().- lwd
(
numeric)
line width. Length of a vector should be equal to number of strata fromsurvival::survfit().- censor_show
(
flag)
whether to show censored.- pch
(
numeric,string)
value or character of points symbol to indicate censored cases.- size
(
numeric)
size of censored point, a class ofunit.- max_time
(
numeric)
maximum value to show on X axis. Only data values less than or up to this threshold value will be plotted (defaults toNULL).- xticks
(
numeric,number, orNULL)
numeric vector of ticks or single number with spacing between ticks on the x axis. IfNULL(default),labeling::extended()is used to determine an optimal tick position on the x axis.- xlab
(
string)
label of x-axis.- yval
(
string)
value of y-axis. Options areSurvival(default) andFailureprobability.- ylab
(
string)
label of y-axis.- ylim
(
vectorofnumeric)
vector of length 2 containing lower and upper limits for the y-axis. IfNULL(default), the minimum and maximum y-values displayed are used as limits.- title
(
string)
title for plot.- footnotes
(
string)
footnotes for plot.- draw
(
flag)
whether the plot should be drawn.- newpage
(
flag)
whether the plot should be drawn on a new page. Only considered ifdraw = TRUEis used.- gp
A
"gpar"object, typically the output from a call to the functiongpar. This is basically a list of graphical parameter settings.- vp
a
viewportobject (orNULL).- name
a character identifier for the grob. Used to find the grob on the display list and/or as a child of another grob.
- font_size
(
number)
font size to be used.- ci_ribbon
(
flag)
draw the confidence interval around the Kaplan-Meier curve.- ggtheme
(
theme)
a graphical theme as provided byggplot2to control outlook of the Kaplan-Meier curve.- annot_at_risk
(
flag)
compute and add the annotation table reporting the number of patient at risk matching the main grid of the Kaplan-Meier curve.- annot_at_risk_title
(
flag)
whether the "Patients at Risk" title should be added above theannot_at_risktable. Has no effect ifannot_at_riskisFALSE. Defaults toTRUE.- annot_surv_med
(
flag)
compute and add the annotation table on the Kaplan-Meier curve estimating the median survival time per group.- annot_coxph
(
flag)
add the annotation table from asurvival::coxph()model.- annot_stats
(
string)
statistics annotations to add to the plot. Options aremedian(median survival follow-up time) andmin(minimum survival follow-up time).- annot_stats_vlines
(
flag)
add vertical lines corresponding to each of the statistics specified byannot_stats. Ifannot_statsisNULLno lines will be added.- control_coxph_pw
-
(
list)
parameters for comparison details, specified by using the helper functioncontrol_coxph(). Some possible parameter options are:pval_method(string)
p-value method for testing hazard ratio = 1. Default method is"log-rank", can also be set to"wald"or"likelihood".ties(string)
method for tie handling. Default is"efron", can also be set to"breslow"or"exact". See more insurvival::coxph()conf_level(proportion)
confidence level of the interval for HR.
- position_coxph
(
numeric)
x and y positions for plottingsurvival::coxph()model.- position_surv_med
(
numeric)
x and y positions for plotting annotation table estimating median survival time per group.- width_annots
(named
listofunits)
a named list of widths for annotation tables with namessurv_med(median survival time table) andcoxph(survival::coxph()model table), where each value is the width (in units) to implement when printing the annotation table.
Examples
# \donttest{
library(dplyr)
library(ggplot2)
library(survival)
library(grid)
library(nestcolor)
df <- tern_ex_adtte %>%
filter(PARAMCD == "OS") %>%
mutate(is_event = CNSR == 0)
variables <- list(tte = "AVAL", is_event = "is_event", arm = "ARMCD")
# 1. Example - basic option
res <- g_km(df = df, variables = variables)
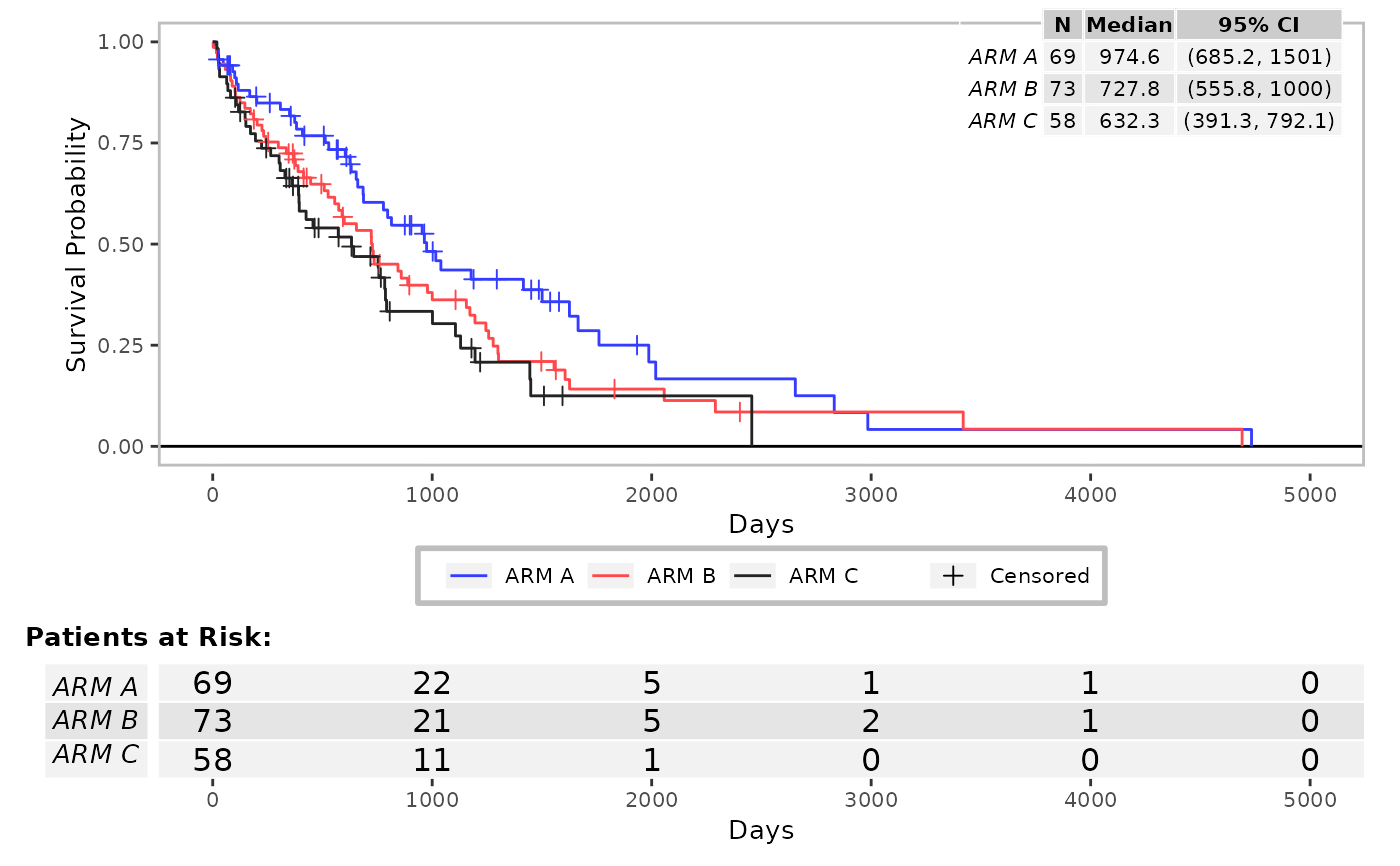 res <- g_km(df = df, variables = variables, yval = "Failure")
res <- g_km(df = df, variables = variables, yval = "Failure")
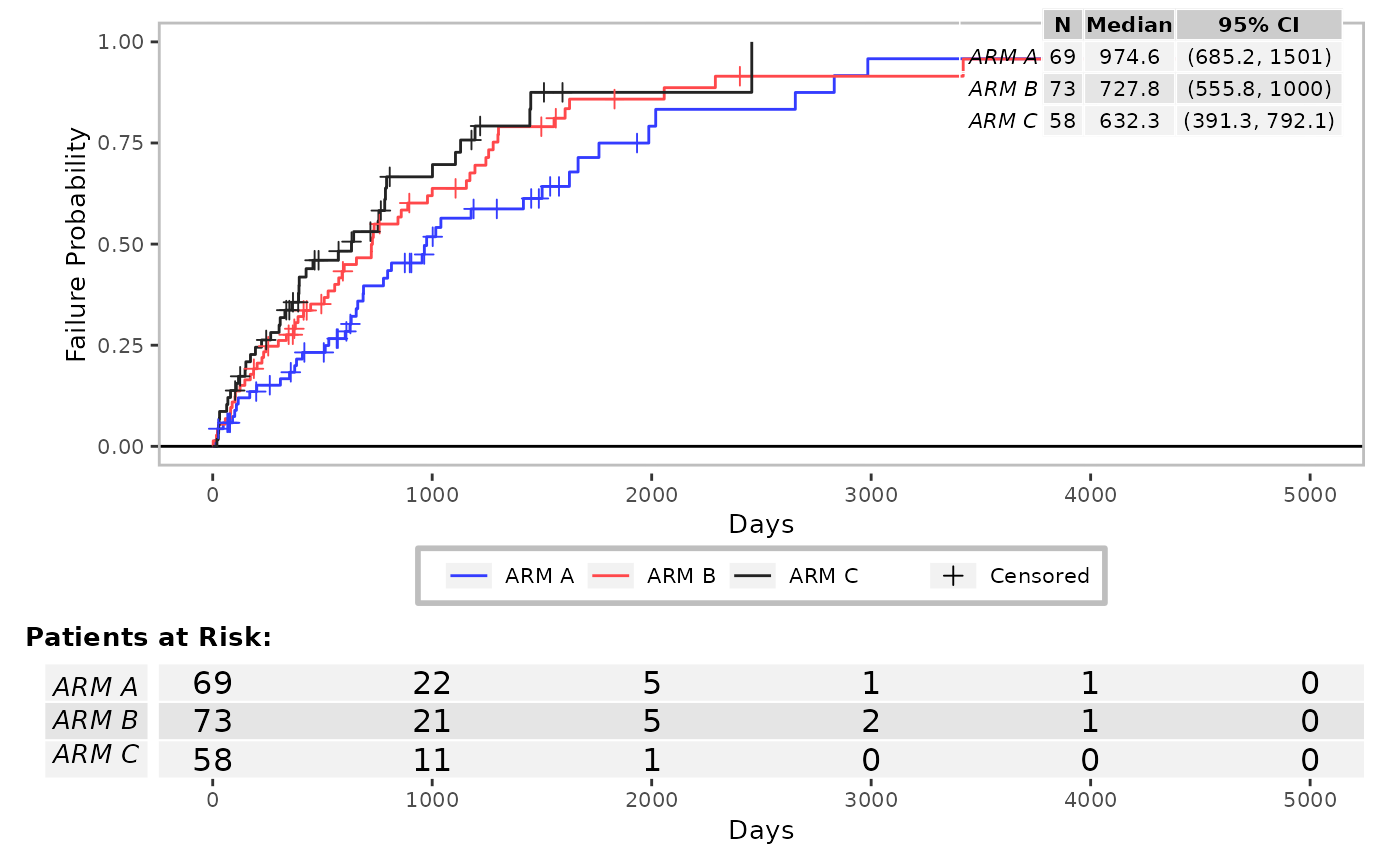 res <- g_km(
df = df,
variables = variables,
control_surv = control_surv_timepoint(conf_level = 0.9),
col = c("grey25", "grey50", "grey75"),
annot_at_risk_title = FALSE
)
res <- g_km(
df = df,
variables = variables,
control_surv = control_surv_timepoint(conf_level = 0.9),
col = c("grey25", "grey50", "grey75"),
annot_at_risk_title = FALSE
)
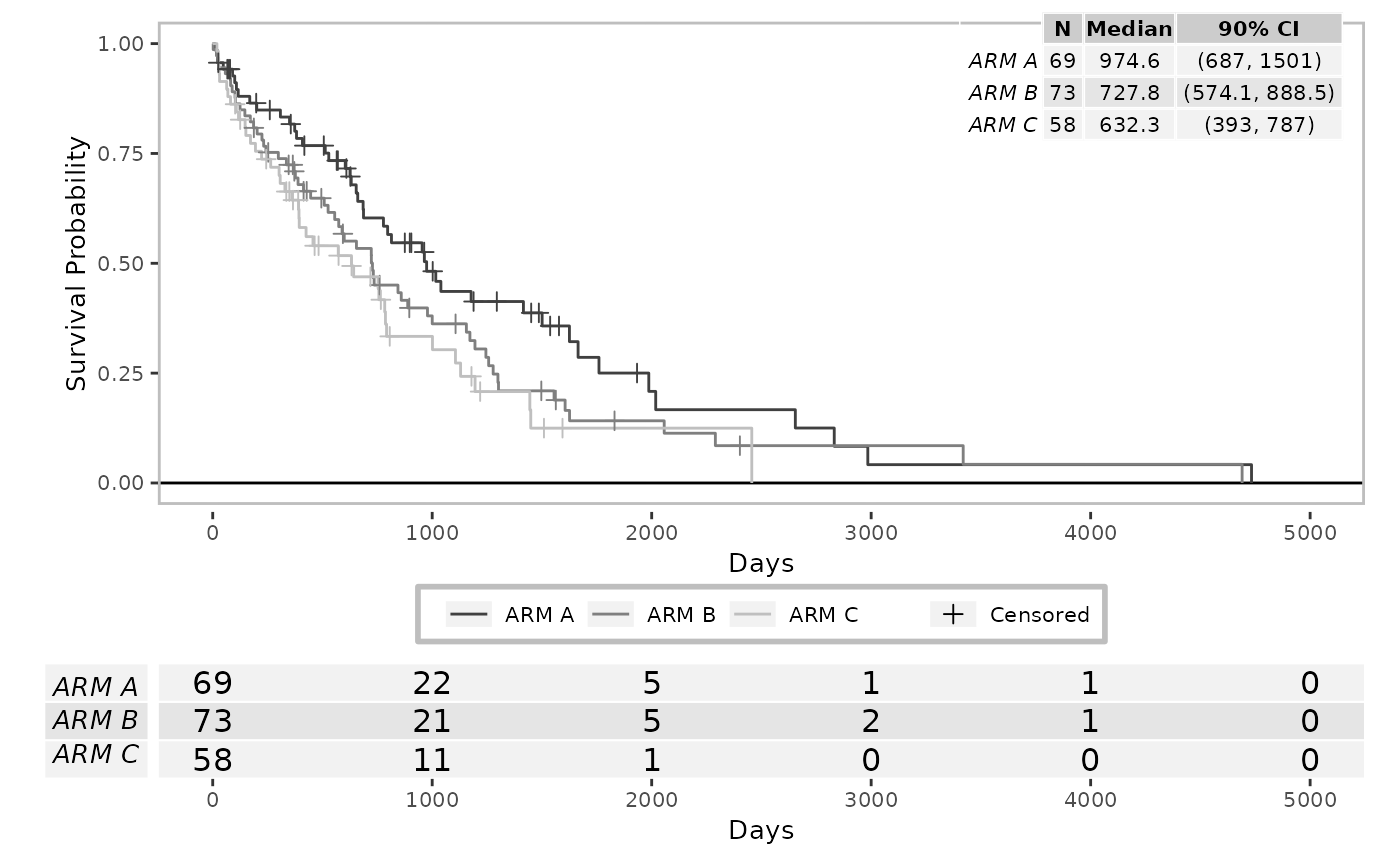 res <- g_km(df = df, variables = variables, ggtheme = theme_minimal())
res <- g_km(df = df, variables = variables, ggtheme = theme_minimal())
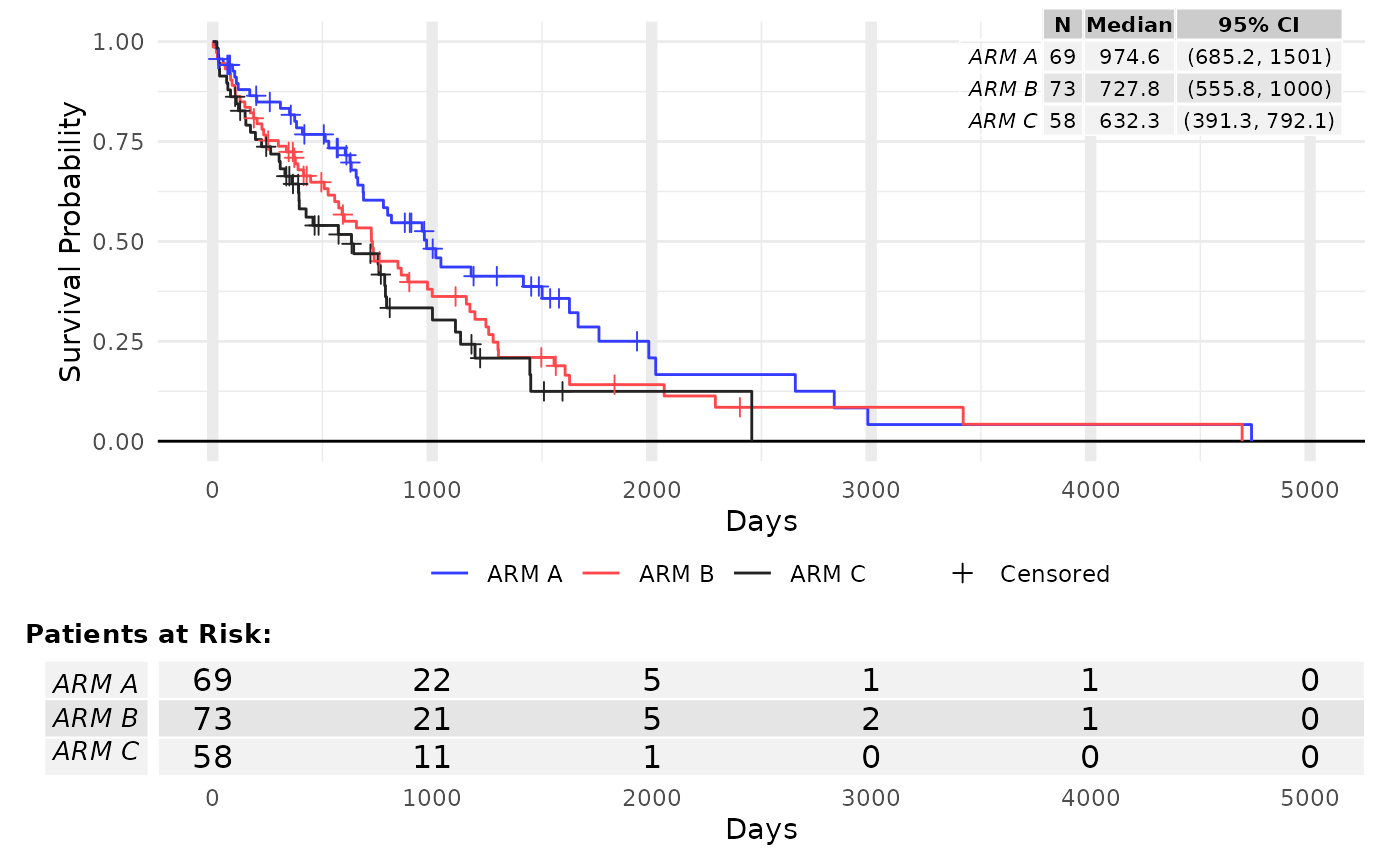 res <- g_km(df = df, variables = variables, ggtheme = theme_minimal(), lty = 1:3)
res <- g_km(df = df, variables = variables, ggtheme = theme_minimal(), lty = 1:3)
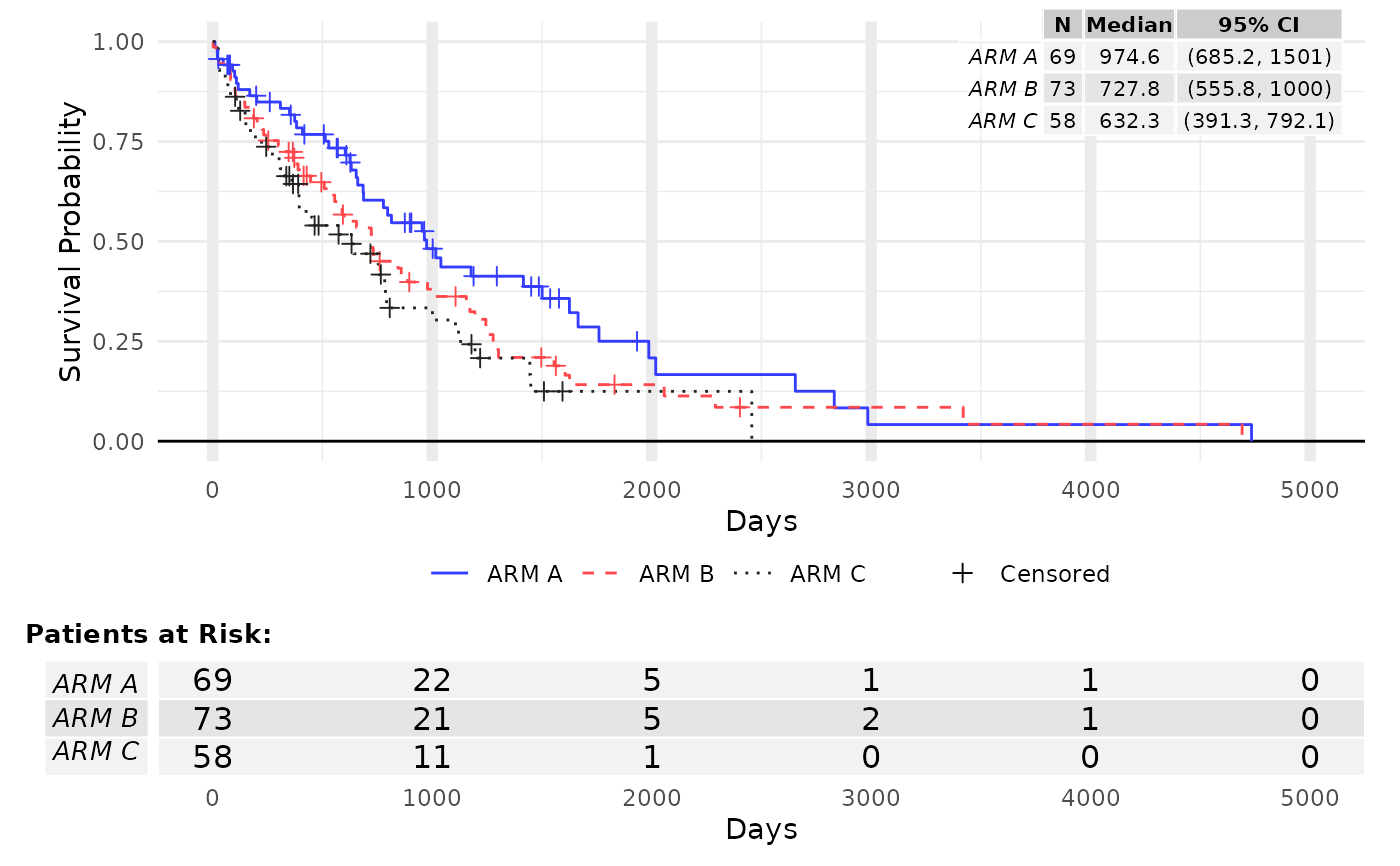 res <- g_km(df = df, variables = variables, max = 2000)
res <- g_km(df = df, variables = variables, max = 2000)
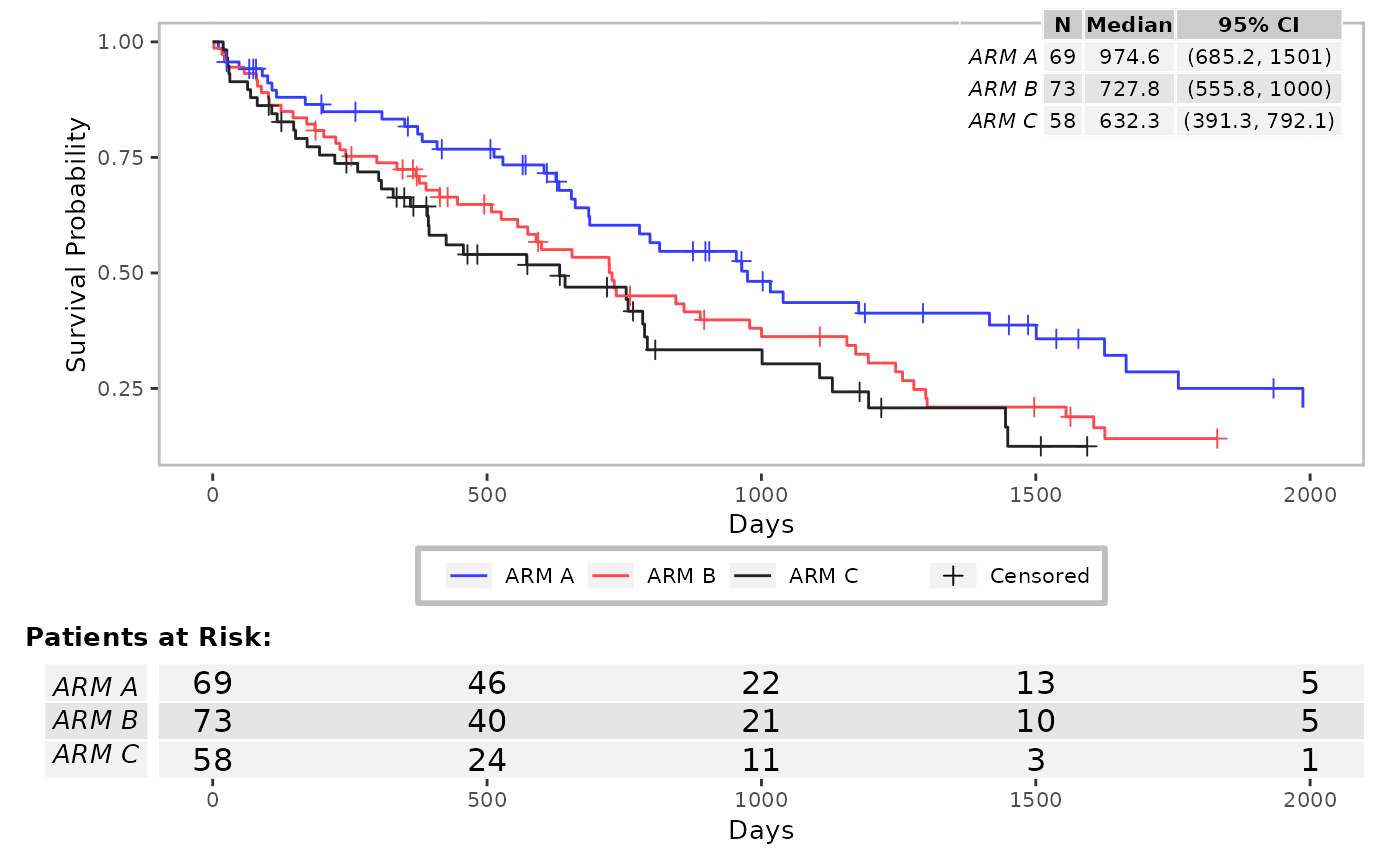 res <- g_km(
df = df,
variables = variables,
annot_stats = c("min", "median"),
annot_stats_vlines = TRUE
)
res <- g_km(
df = df,
variables = variables,
annot_stats = c("min", "median"),
annot_stats_vlines = TRUE
)
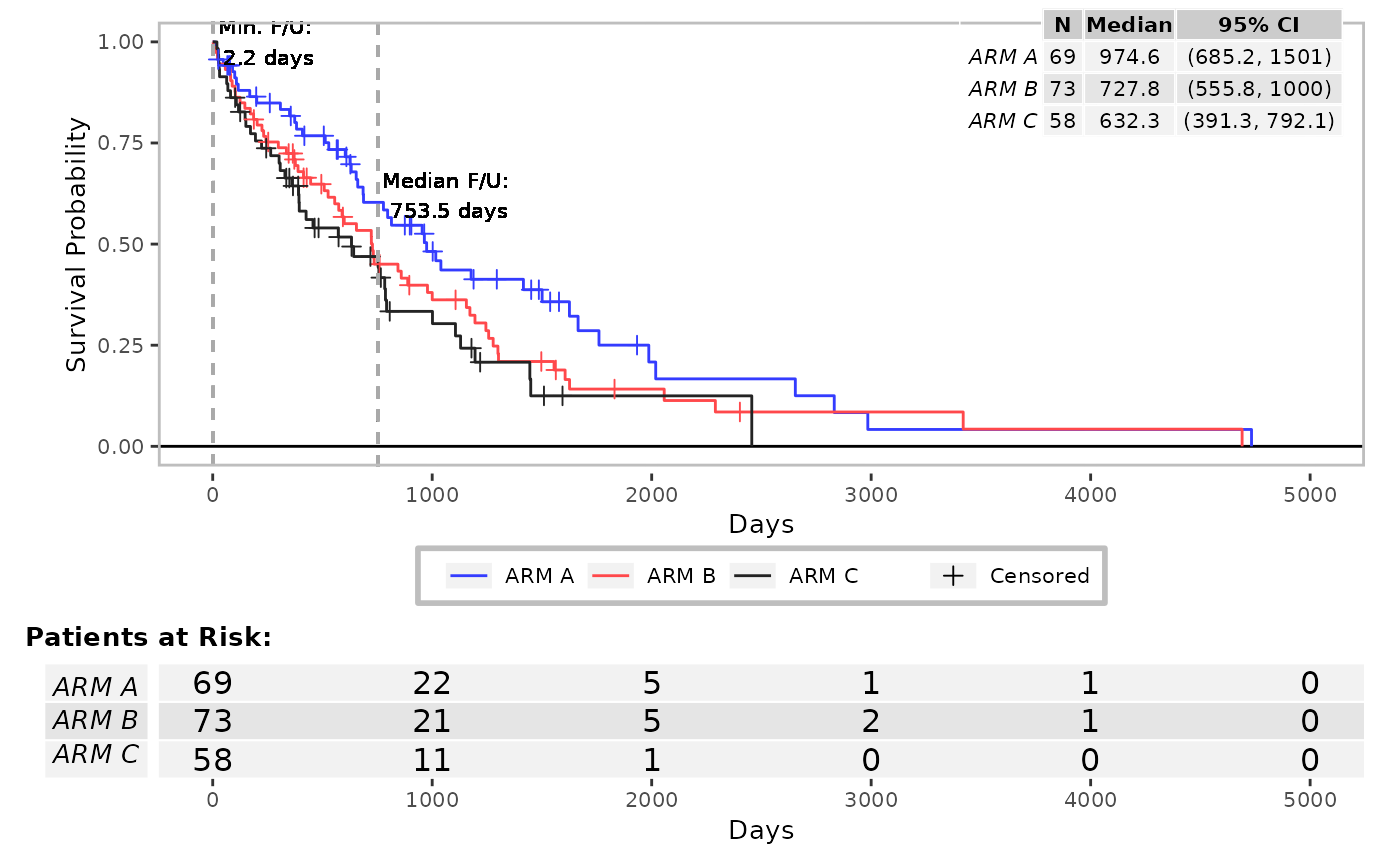 # 2. Example - Arrange several KM curve on a single graph device
# 2.1 Use case: A general graph on the top, a zoom on the bottom.
grid.newpage()
lyt <- grid.layout(nrow = 2, ncol = 1) %>%
viewport(layout = .) %>%
pushViewport()
res <- g_km(
df = df, variables = variables, newpage = FALSE, annot_surv_med = FALSE,
vp = viewport(layout.pos.row = 1, layout.pos.col = 1)
)
res <- g_km(
df = df, variables = variables, max = 1000, newpage = FALSE, annot_surv_med = FALSE,
ggtheme = theme_dark(),
vp = viewport(layout.pos.row = 2, layout.pos.col = 1)
)
# 2. Example - Arrange several KM curve on a single graph device
# 2.1 Use case: A general graph on the top, a zoom on the bottom.
grid.newpage()
lyt <- grid.layout(nrow = 2, ncol = 1) %>%
viewport(layout = .) %>%
pushViewport()
res <- g_km(
df = df, variables = variables, newpage = FALSE, annot_surv_med = FALSE,
vp = viewport(layout.pos.row = 1, layout.pos.col = 1)
)
res <- g_km(
df = df, variables = variables, max = 1000, newpage = FALSE, annot_surv_med = FALSE,
ggtheme = theme_dark(),
vp = viewport(layout.pos.row = 2, layout.pos.col = 1)
)
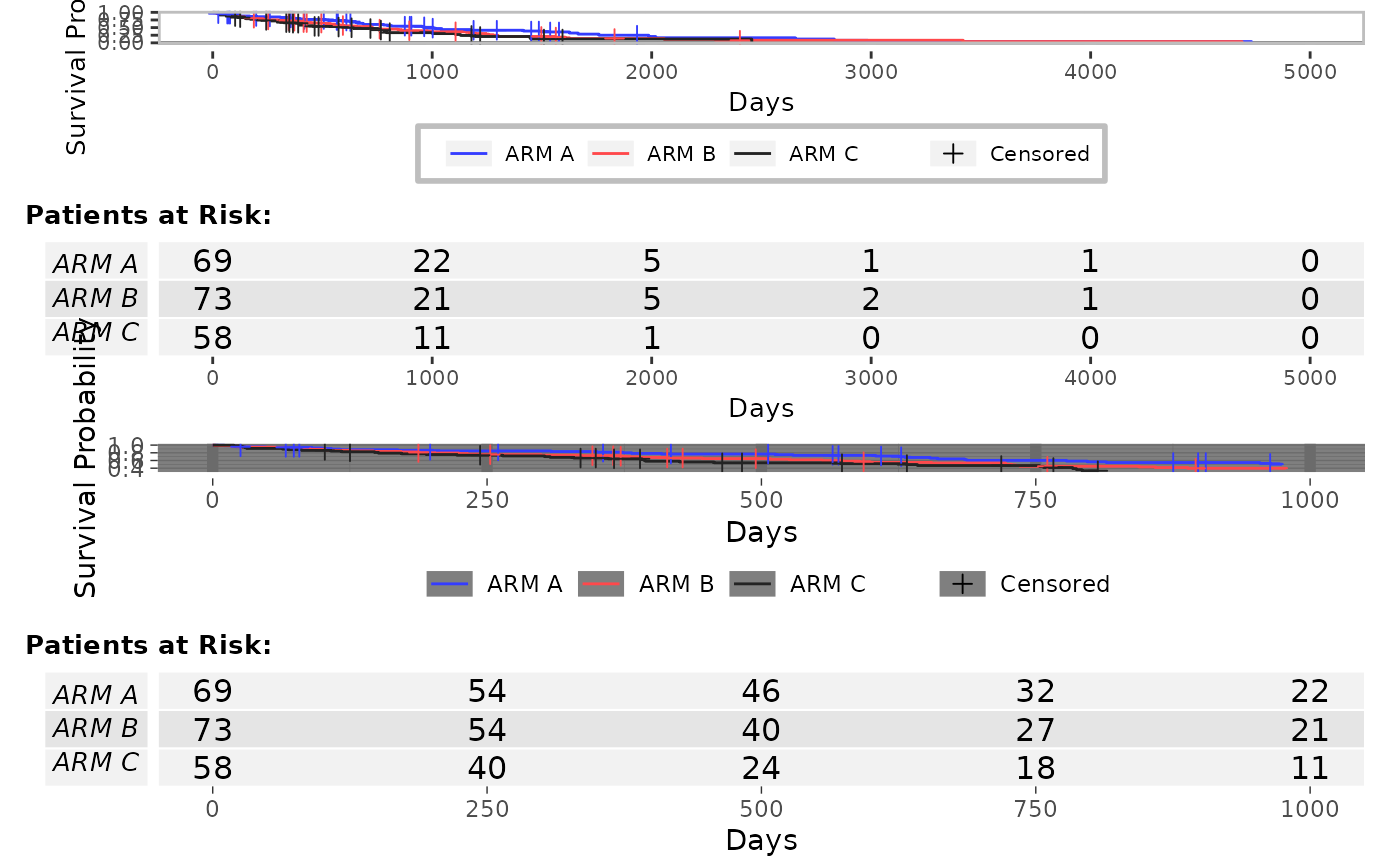 # 2.1 Use case: No annotations on top, annotated graph on bottom
grid.newpage()
lyt <- grid.layout(nrow = 2, ncol = 1) %>%
viewport(layout = .) %>%
pushViewport()
res <- g_km(
df = df, variables = variables, newpage = FALSE,
annot_surv_med = FALSE, annot_at_risk = FALSE,
vp = viewport(layout.pos.row = 1, layout.pos.col = 1)
)
res <- g_km(
df = df, variables = variables, max = 2000, newpage = FALSE, annot_surv_med = FALSE,
annot_at_risk = TRUE,
ggtheme = theme_dark(),
vp = viewport(layout.pos.row = 2, layout.pos.col = 1)
)
# 2.1 Use case: No annotations on top, annotated graph on bottom
grid.newpage()
lyt <- grid.layout(nrow = 2, ncol = 1) %>%
viewport(layout = .) %>%
pushViewport()
res <- g_km(
df = df, variables = variables, newpage = FALSE,
annot_surv_med = FALSE, annot_at_risk = FALSE,
vp = viewport(layout.pos.row = 1, layout.pos.col = 1)
)
res <- g_km(
df = df, variables = variables, max = 2000, newpage = FALSE, annot_surv_med = FALSE,
annot_at_risk = TRUE,
ggtheme = theme_dark(),
vp = viewport(layout.pos.row = 2, layout.pos.col = 1)
)
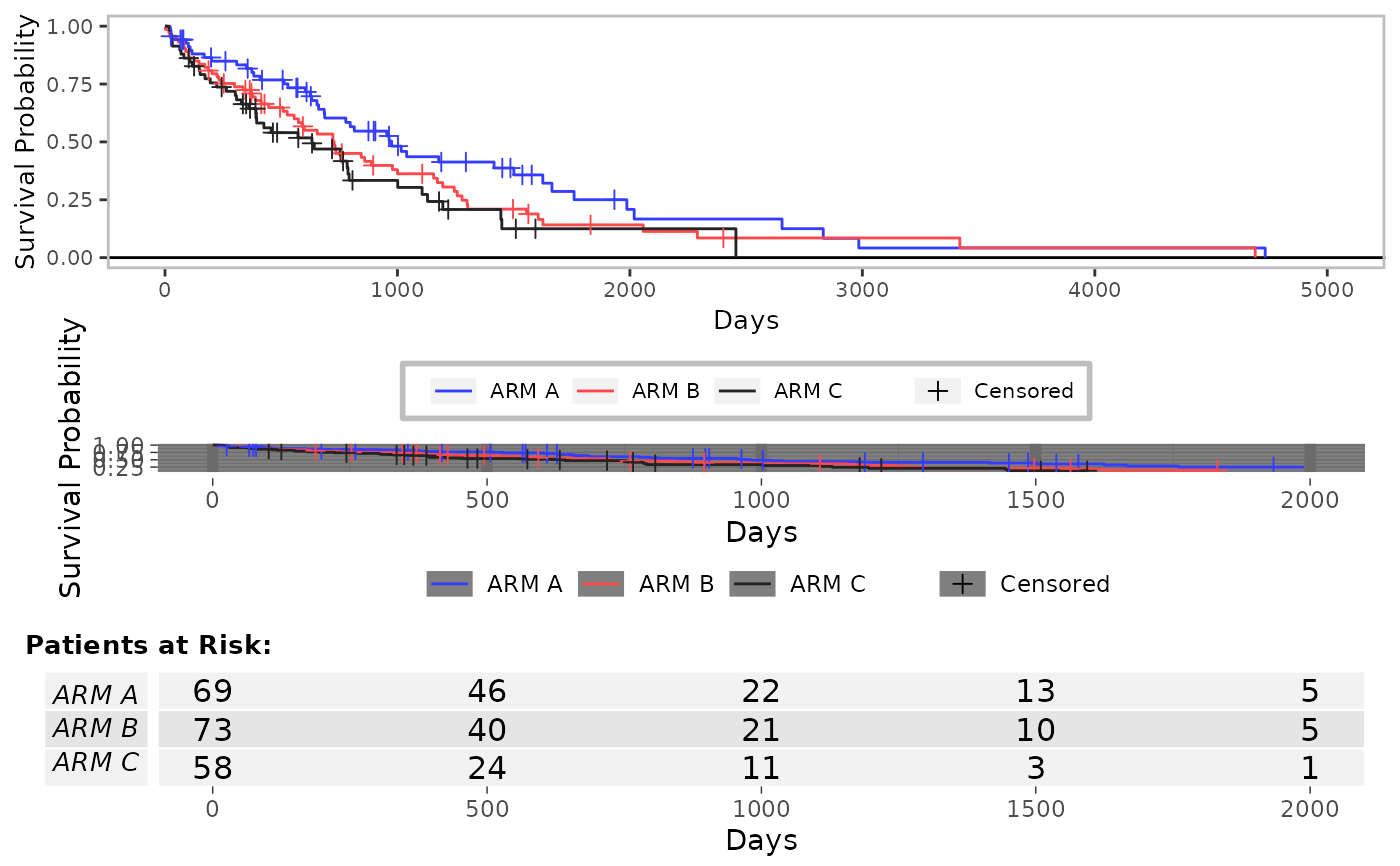 # Add annotation from a pairwise coxph analysis
g_km(
df = df, variables = variables,
annot_coxph = TRUE
)
# Add annotation from a pairwise coxph analysis
g_km(
df = df, variables = variables,
annot_coxph = TRUE
)
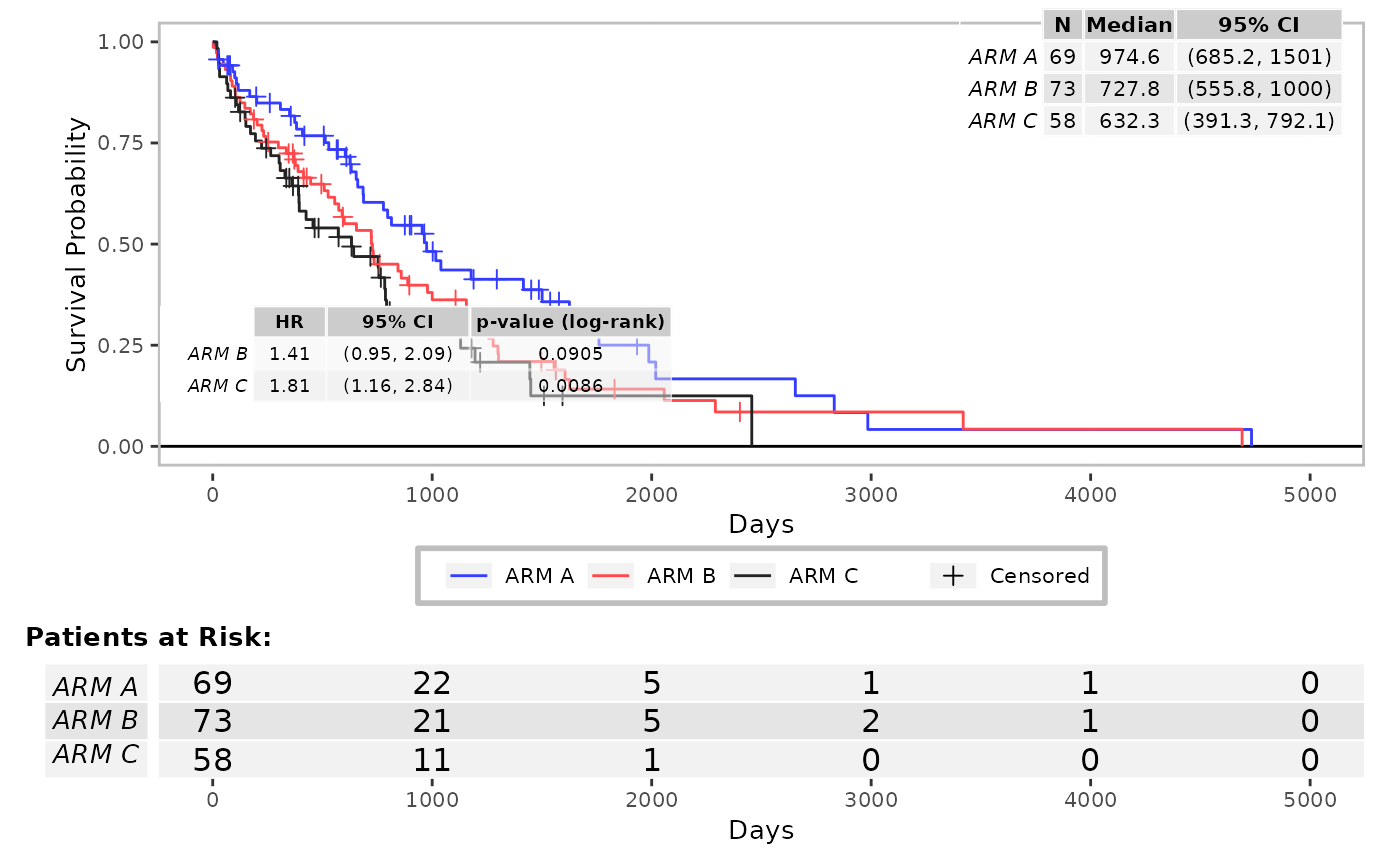 # Change widths/sizes of surv_med and coxph annotation tables.
g_km(
df = df, variables = c(variables, list(strat = "SEX")),
annot_coxph = TRUE,
width_annots = list(surv_med = grid::unit(2, "in"), coxph = grid::unit(3, "in"))
)
# Change widths/sizes of surv_med and coxph annotation tables.
g_km(
df = df, variables = c(variables, list(strat = "SEX")),
annot_coxph = TRUE,
width_annots = list(surv_med = grid::unit(2, "in"), coxph = grid::unit(3, "in"))
)
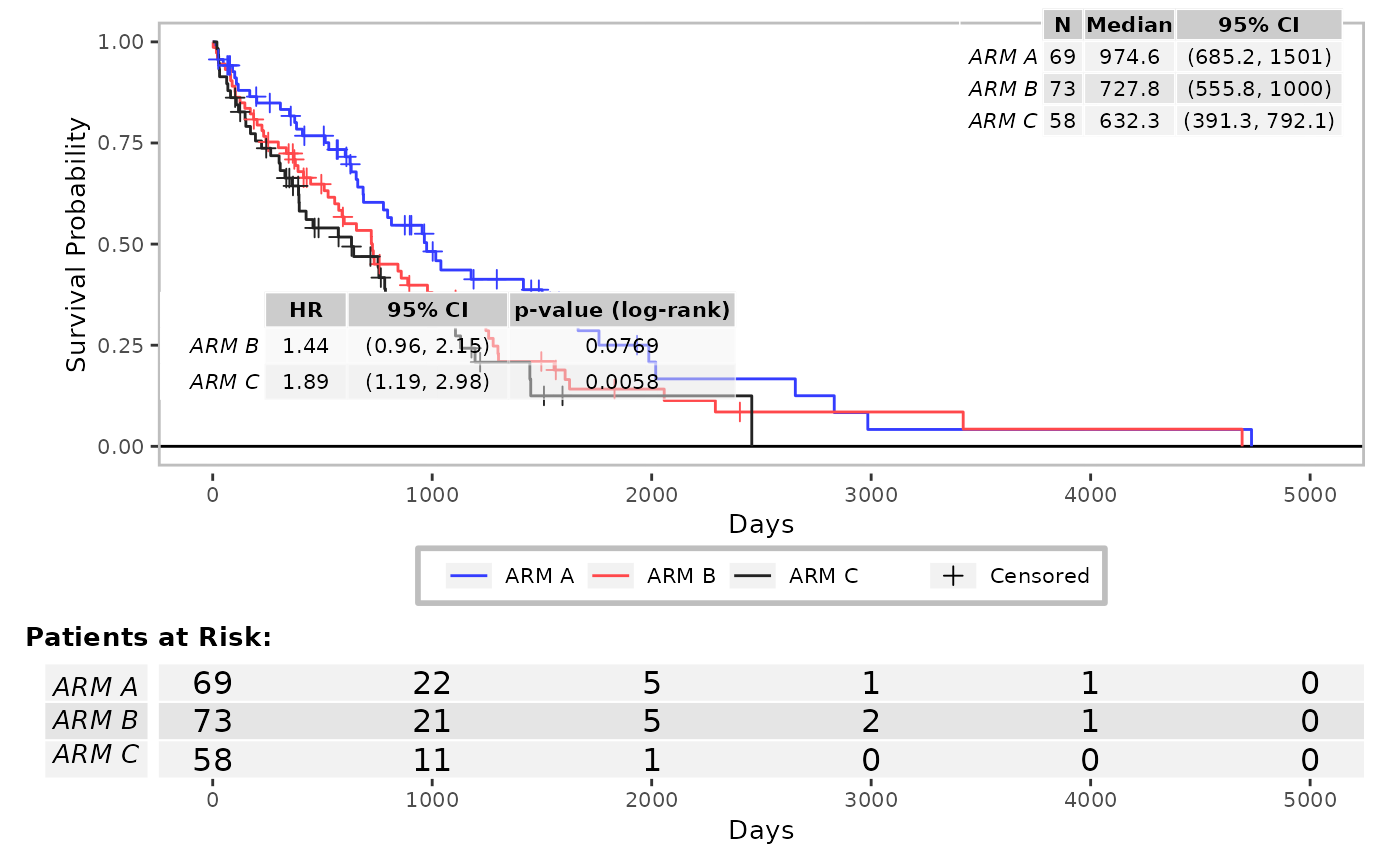 g_km(
df = df, variables = c(variables, list(strat = "SEX")),
font_size = 15,
annot_coxph = TRUE,
control_coxph = control_coxph(pval_method = "wald", ties = "exact", conf_level = 0.99),
position_coxph = c(0.5, 0.5)
)
g_km(
df = df, variables = c(variables, list(strat = "SEX")),
font_size = 15,
annot_coxph = TRUE,
control_coxph = control_coxph(pval_method = "wald", ties = "exact", conf_level = 0.99),
position_coxph = c(0.5, 0.5)
)
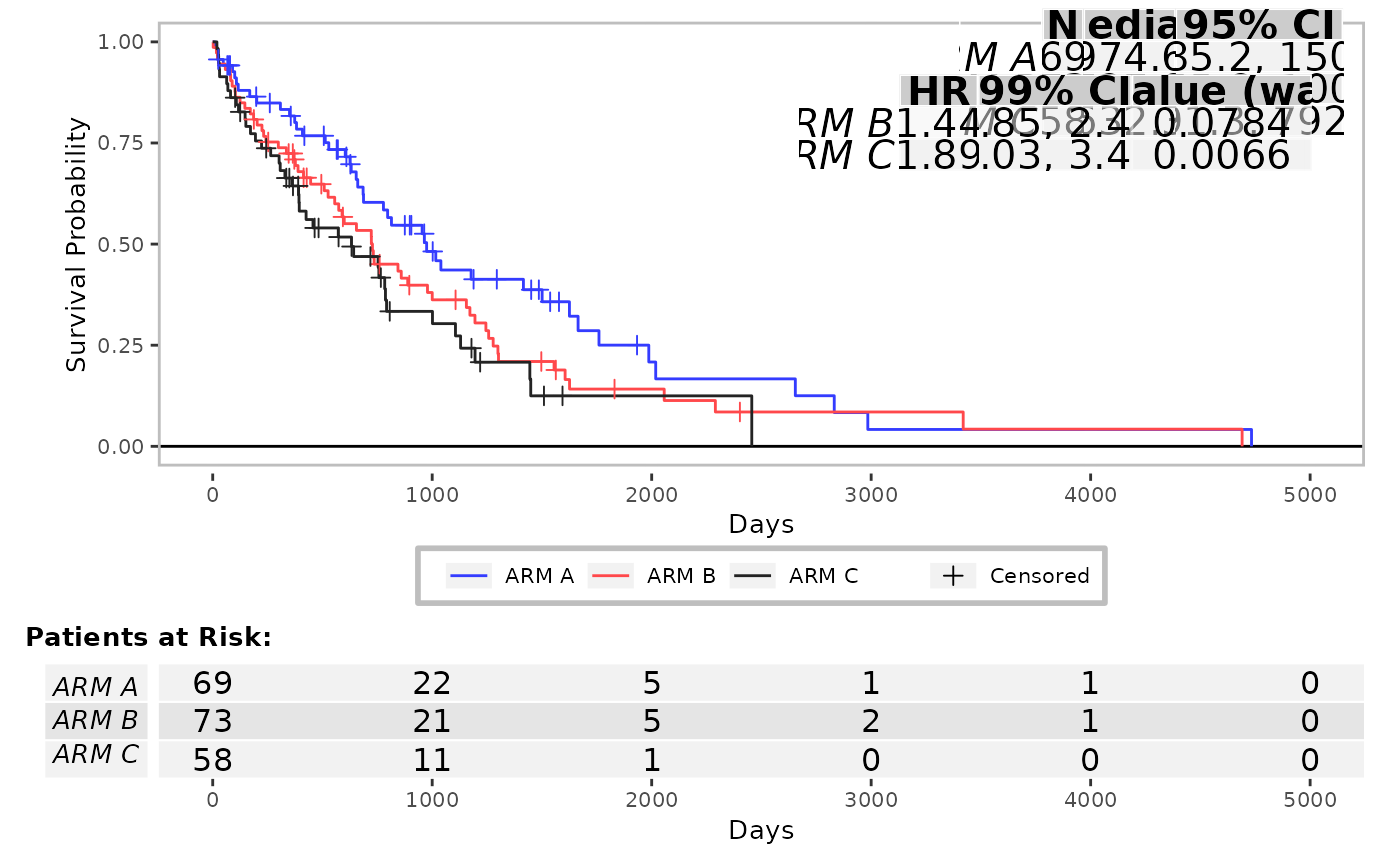 # Change position of the treatment group annotation table.
g_km(
df = df, variables = c(variables, list(strat = "SEX")),
font_size = 15,
annot_coxph = TRUE,
control_coxph = control_coxph(pval_method = "wald", ties = "exact", conf_level = 0.99),
position_surv_med = c(1, 0.7)
)
# Change position of the treatment group annotation table.
g_km(
df = df, variables = c(variables, list(strat = "SEX")),
font_size = 15,
annot_coxph = TRUE,
control_coxph = control_coxph(pval_method = "wald", ties = "exact", conf_level = 0.99),
position_surv_med = c(1, 0.7)
)
 # }
# }