Usage
g_forest(
tbl,
col_x = attr(tbl, "col_x"),
col_ci = attr(tbl, "col_ci"),
vline = 1,
forest_header = attr(tbl, "forest_header"),
xlim = c(0.1, 10),
logx = TRUE,
x_at = c(0.1, 1, 10),
width_row_names = NULL,
width_columns = NULL,
width_forest = grid::unit(1, "null"),
col_symbol_size = attr(tbl, "col_symbol_size"),
col = getOption("ggplot2.discrete.colour")[1],
draw = TRUE,
newpage = TRUE
)Arguments
- tbl
(
rtable)- col_x
(
integer)
column index with estimator. By default tries to get this fromtblattributecol_x, otherwise needs to be manually specified.- col_ci
(
integer)
column index with confidence intervals. By default tries to get this fromtblattributecol_ci, otherwise needs to be manually specified.- vline
(
number)
x coordinate for vertical line, ifNULLthen the line is omitted.- forest_header
(
character, length 2)
text displayed to the left and right ofvline, respectively. Ifvline = NULLthenforest_headerneeds to beNULLtoo. By default tries to get this fromtblattributeforest_header.- xlim
(
numeric)
limits for x axis.- logx
(
flag)
show the x-values on logarithm scale.- x_at
(
numeric)
x-tick locations, ifNULLthey get automatically chosen.- width_row_names
(
unit)
width for row names. IfNULLthe widths get automatically calculated. Seegrid::unit().- width_columns
(
unit)
widths for the table columns. IfNULLthe widths get automatically calculated. Seegrid::unit().- width_forest
(
unit)
width for the forest column. IfNULLthe widths get automatically calculated. Seegrid::unit().- col_symbol_size
(
integer)
column index fromtblcontaining data to be used to determine relative size for estimator plot symbol. Typically, the symbol size is proportional to the sample size used to calculate the estimator. IfNULL, the same symbol size is used for all subgroups. By default tries to get this fromtblattributecol_symbol_size, otherwise needs to be manually specified.- col
(
character)
color(s).- draw
(
flag)
whether the plot should be drawn.- newpage
(
flag)
whether the plot should be drawn on a new page. Only considered ifdraw = TRUEis used.
Details
Create a forest plot from any rtables::rtable() object that has a
column with a single value and a column with 2 values.
Examples
# \donttest{
library(dplyr)
library(forcats)
library(nestcolor)
adrs <- tern_ex_adrs
n_records <- 20
adrs_labels <- formatters::var_labels(adrs, fill = TRUE)
adrs <- adrs %>%
filter(PARAMCD == "BESRSPI") %>%
filter(ARM %in% c("A: Drug X", "B: Placebo")) %>%
slice(seq_len(n_records)) %>%
droplevels() %>%
mutate(
# Reorder levels of factor to make the placebo group the reference arm.
ARM = fct_relevel(ARM, "B: Placebo"),
rsp = AVALC == "CR"
)
formatters::var_labels(adrs) <- c(adrs_labels, "Response")
df <- extract_rsp_subgroups(
variables = list(rsp = "rsp", arm = "ARM", subgroups = c("SEX", "STRATA2")),
data = adrs
)
# Full commonly used response table.
tbl <- basic_table() %>%
tabulate_rsp_subgroups(df)
p <- g_forest(tbl)
draw_grob(p)
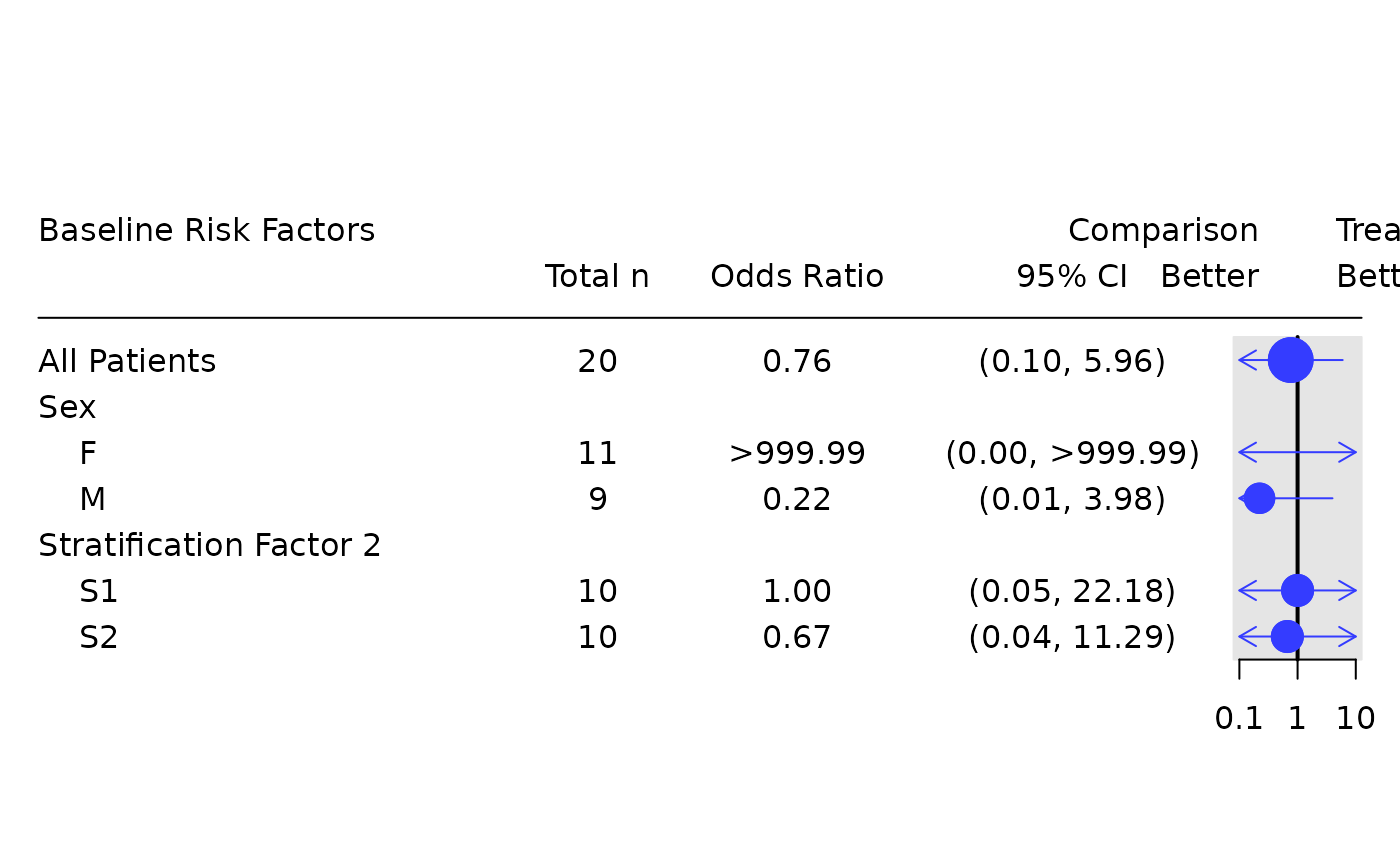
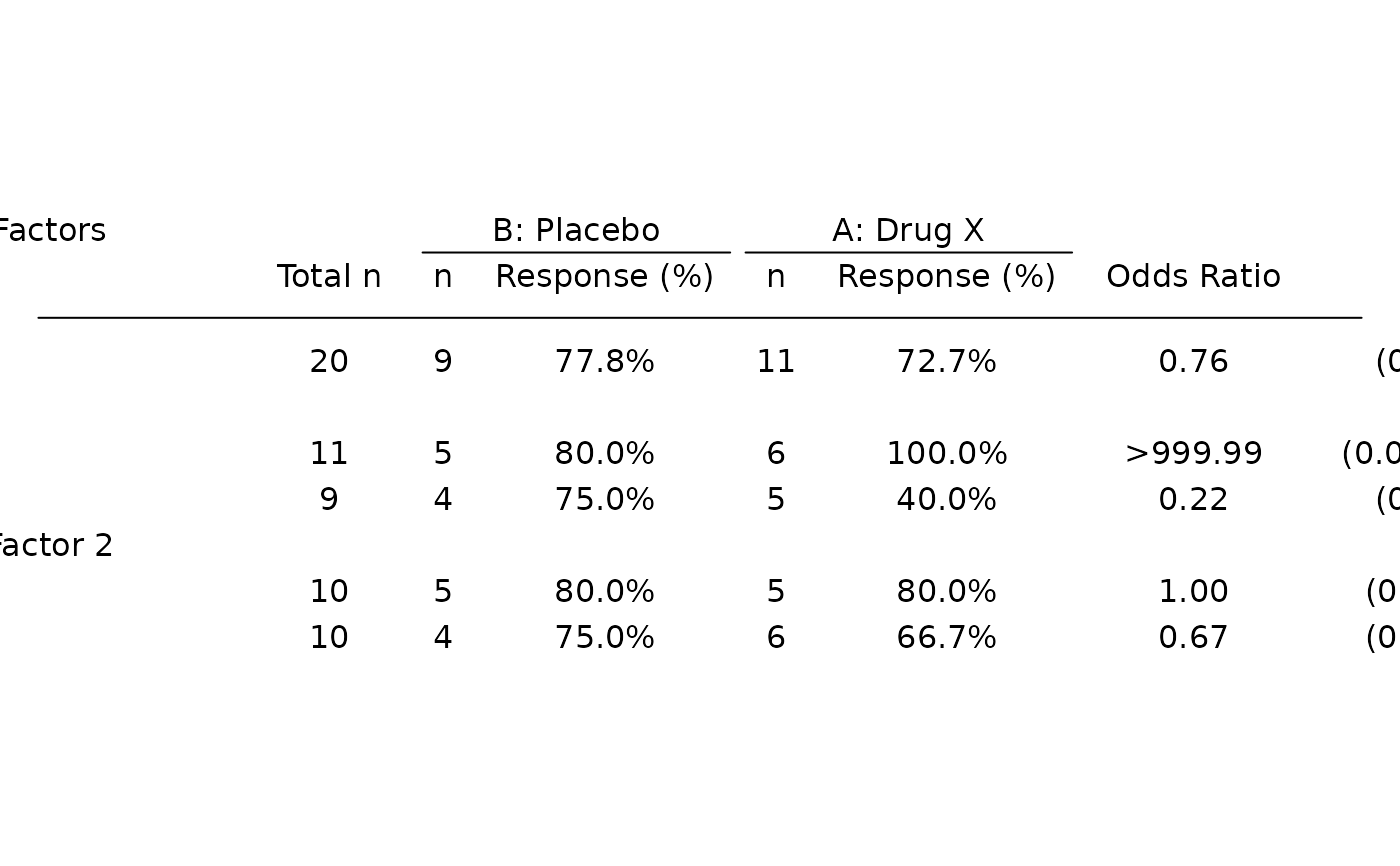 # Odds ratio only table.
tbl_or <- basic_table() %>%
tabulate_rsp_subgroups(df, vars = c("n_tot", "or", "ci"))
tbl_or
#> Baseline Risk Factors
#> Total n Odds Ratio 95% CI
#> ————————————————————————————————————————————————————————————————
#> All Patients 20 0.76 (0.10, 5.96)
#> Sex
#> F 11 >999.99 (0.00, >999.99)
#> M 9 0.22 (0.01, 3.98)
#> Stratification Factor 2
#> S1 10 1.00 (0.05, 22.18)
#> S2 10 0.67 (0.04, 11.29)
p <- g_forest(
tbl_or,
forest_header = c("Comparison\nBetter", "Treatment\nBetter")
)
draw_grob(p)
# Odds ratio only table.
tbl_or <- basic_table() %>%
tabulate_rsp_subgroups(df, vars = c("n_tot", "or", "ci"))
tbl_or
#> Baseline Risk Factors
#> Total n Odds Ratio 95% CI
#> ————————————————————————————————————————————————————————————————
#> All Patients 20 0.76 (0.10, 5.96)
#> Sex
#> F 11 >999.99 (0.00, >999.99)
#> M 9 0.22 (0.01, 3.98)
#> Stratification Factor 2
#> S1 10 1.00 (0.05, 22.18)
#> S2 10 0.67 (0.04, 11.29)
p <- g_forest(
tbl_or,
forest_header = c("Comparison\nBetter", "Treatment\nBetter")
)
draw_grob(p)
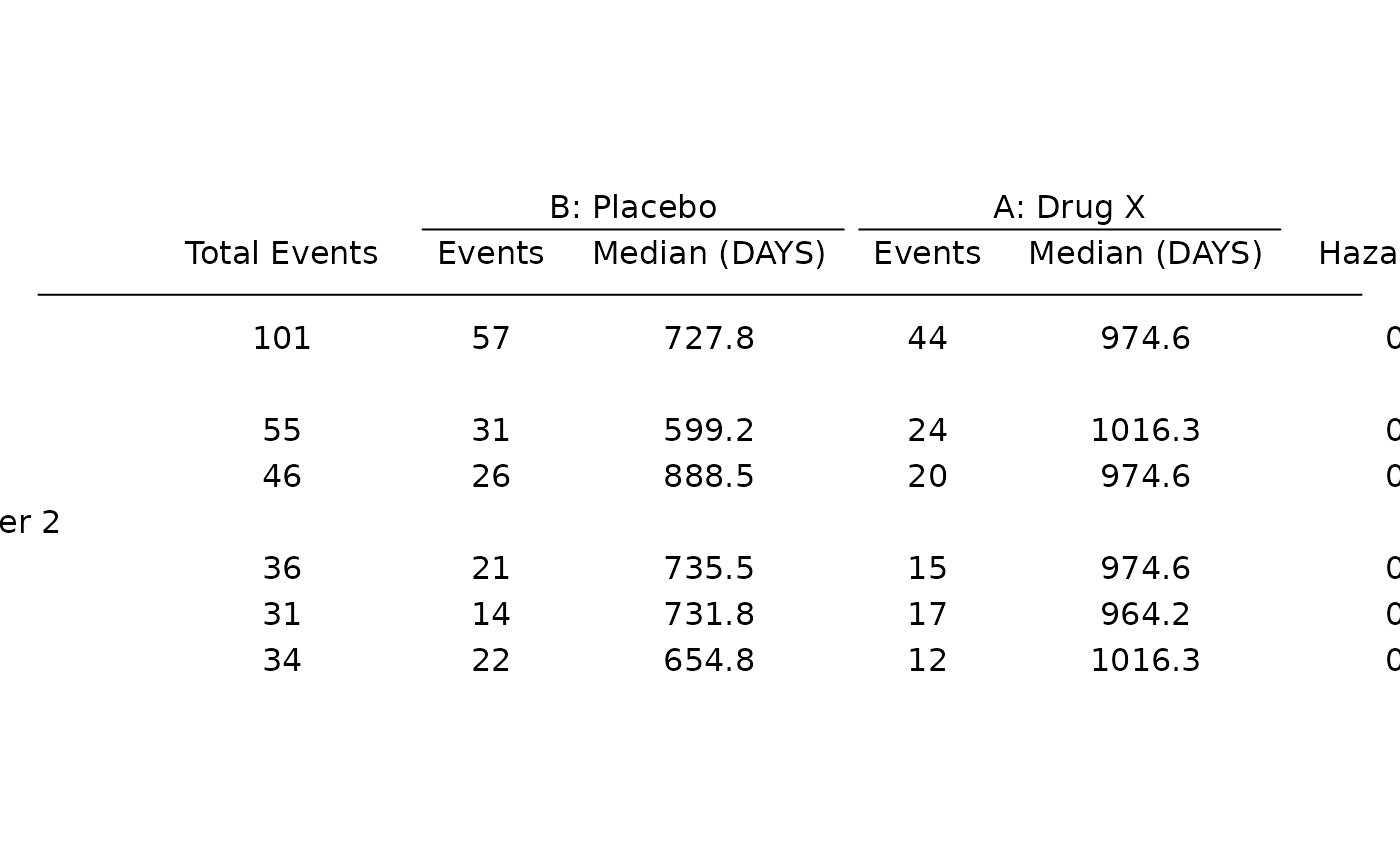
 # Survival forest plot example.
adtte <- tern_ex_adtte
# Save variable labels before data processing steps.
adtte_labels <- formatters::var_labels(adtte, fill = TRUE)
adtte_f <- adtte %>%
filter(
PARAMCD == "OS",
ARM %in% c("B: Placebo", "A: Drug X"),
SEX %in% c("M", "F")
) %>%
mutate(
# Reorder levels of ARM to display reference arm before treatment arm.
ARM = droplevels(fct_relevel(ARM, "B: Placebo")),
SEX = droplevels(SEX),
AVALU = as.character(AVALU),
is_event = CNSR == 0
)
labels <- list(
"ARM" = adtte_labels["ARM"],
"SEX" = adtte_labels["SEX"],
"AVALU" = adtte_labels["AVALU"],
"is_event" = "Event Flag"
)
formatters::var_labels(adtte_f)[names(labels)] <- as.character(labels)
df <- extract_survival_subgroups(
variables = list(
tte = "AVAL",
is_event = "is_event",
arm = "ARM", subgroups = c("SEX", "BMRKR2")
),
data = adtte_f
)
table_hr <- basic_table() %>%
tabulate_survival_subgroups(df, time_unit = adtte_f$AVALU[1])
g_forest(table_hr)
# Survival forest plot example.
adtte <- tern_ex_adtte
# Save variable labels before data processing steps.
adtte_labels <- formatters::var_labels(adtte, fill = TRUE)
adtte_f <- adtte %>%
filter(
PARAMCD == "OS",
ARM %in% c("B: Placebo", "A: Drug X"),
SEX %in% c("M", "F")
) %>%
mutate(
# Reorder levels of ARM to display reference arm before treatment arm.
ARM = droplevels(fct_relevel(ARM, "B: Placebo")),
SEX = droplevels(SEX),
AVALU = as.character(AVALU),
is_event = CNSR == 0
)
labels <- list(
"ARM" = adtte_labels["ARM"],
"SEX" = adtte_labels["SEX"],
"AVALU" = adtte_labels["AVALU"],
"is_event" = "Event Flag"
)
formatters::var_labels(adtte_f)[names(labels)] <- as.character(labels)
df <- extract_survival_subgroups(
variables = list(
tte = "AVAL",
is_event = "is_event",
arm = "ARM", subgroups = c("SEX", "BMRKR2")
),
data = adtte_f
)
table_hr <- basic_table() %>%
tabulate_survival_subgroups(df, time_unit = adtte_f$AVALU[1])
g_forest(table_hr)
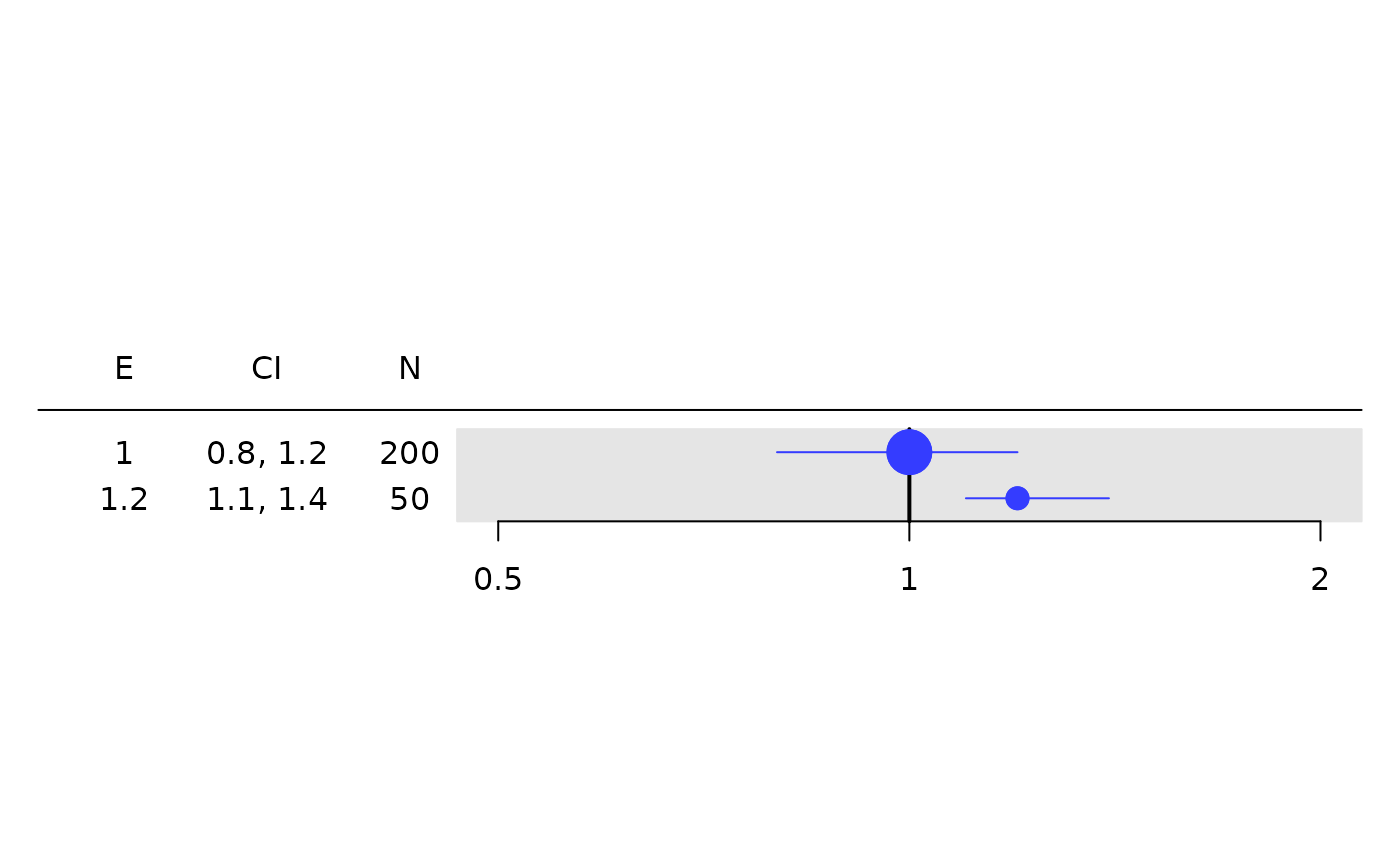
 # Works with any `rtable`.
tbl <- rtable(
header = c("E", "CI", "N"),
rrow("", 1, c(.8, 1.2), 200),
rrow("", 1.2, c(1.1, 1.4), 50)
)
g_forest(
tbl = tbl,
col_x = 1,
col_ci = 2,
xlim = c(0.5, 2),
x_at = c(0.5, 1, 2),
col_symbol_size = 3
)
# Works with any `rtable`.
tbl <- rtable(
header = c("E", "CI", "N"),
rrow("", 1, c(.8, 1.2), 200),
rrow("", 1.2, c(1.1, 1.4), 50)
)
g_forest(
tbl = tbl,
col_x = 1,
col_ci = 2,
xlim = c(0.5, 2),
x_at = c(0.5, 1, 2),
col_symbol_size = 3
)
 tbl <- rtable(
header = rheader(
rrow("", rcell("A", colspan = 2)),
rrow("", "c1", "c2")
),
rrow("row 1", 1, c(.8, 1.2)),
rrow("row 2", 1.2, c(1.1, 1.4))
)
g_forest(
tbl = tbl,
col_x = 1,
col_ci = 2,
xlim = c(0.5, 2),
x_at = c(0.5, 1, 2),
vline = 1,
forest_header = c("Hello", "World")
)
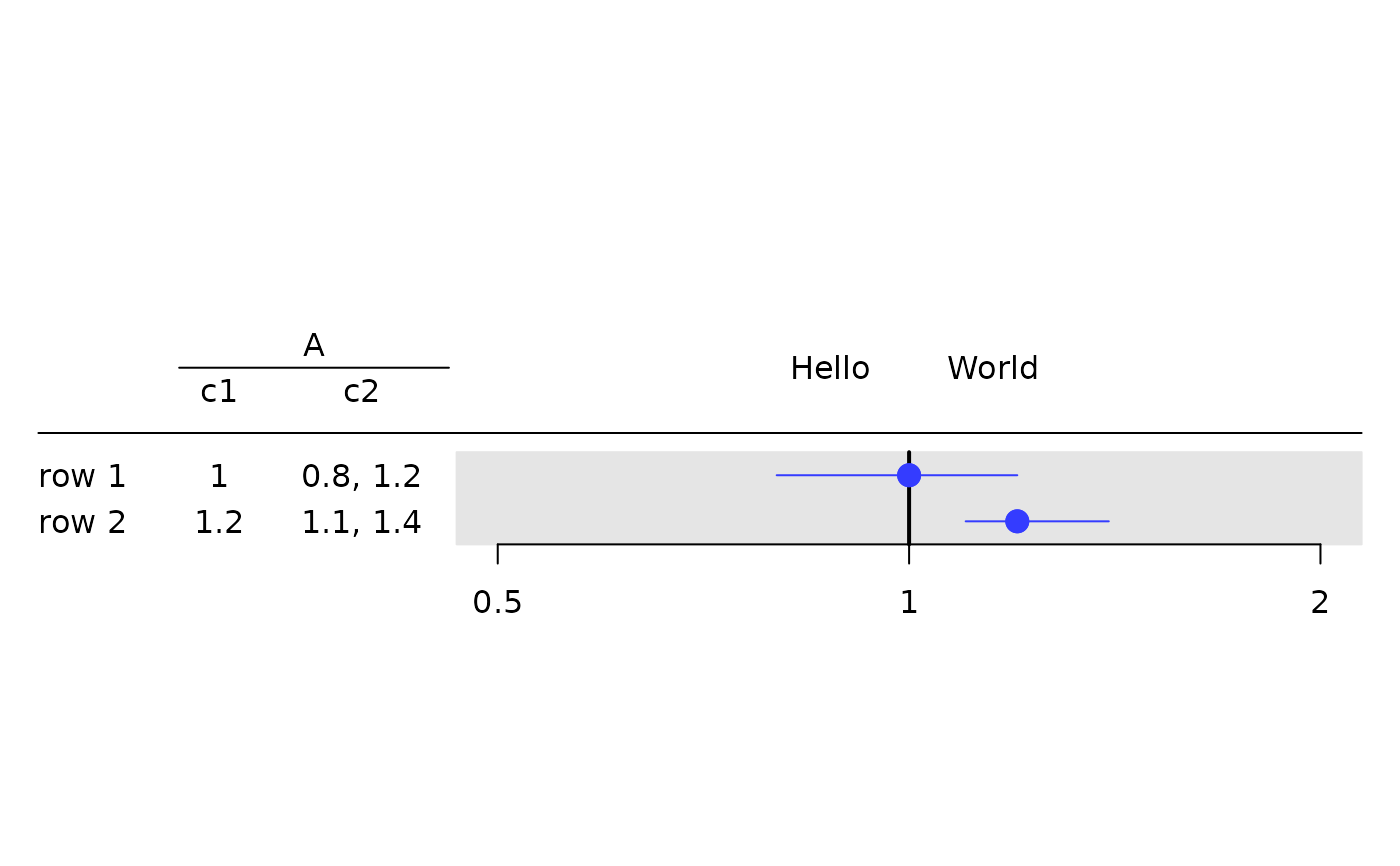
tbl <- rtable(
header = rheader(
rrow("", rcell("A", colspan = 2)),
rrow("", "c1", "c2")
),
rrow("row 1", 1, c(.8, 1.2)),
rrow("row 2", 1.2, c(1.1, 1.4))
)
g_forest(
tbl = tbl,
col_x = 1,
col_ci = 2,
xlim = c(0.5, 2),
x_at = c(0.5, 1, 2),
vline = 1,
forest_header = c("Hello", "World")
)
 # }
# }